Resources
Nanopore-based classification of acute leukemia
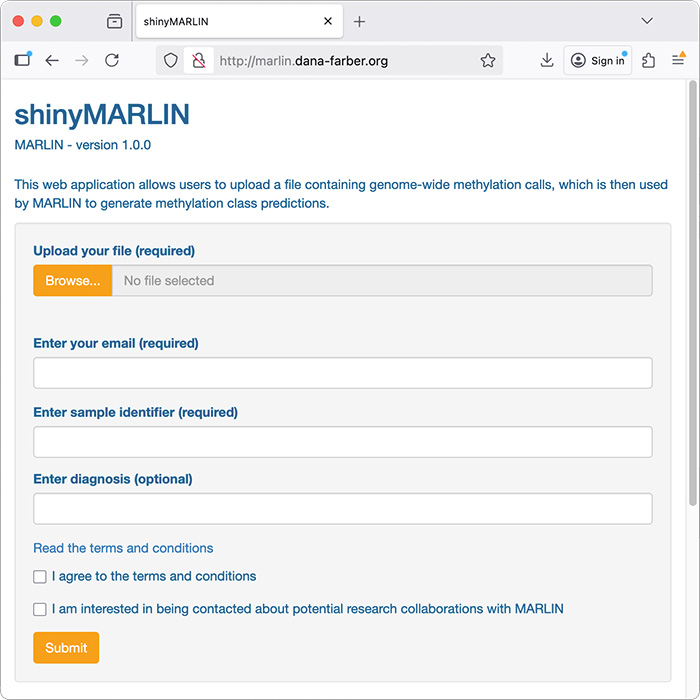
Web implementation of MARLIN, our machine learning classifier for acute leukemia. MARLIN (Methylation- and AI-guided Rapid Leukemia Subtype Inference) is compatible with different DNA methylation profiling technologies, including low-coverage nanopore sequencing, and generates class predictions within minutes. Please also see the GitHub repository for the trained model and scripts.
http://marlin.dana-farber.org/Explainable AI of DNA methylation-based brain tumor diagnostics
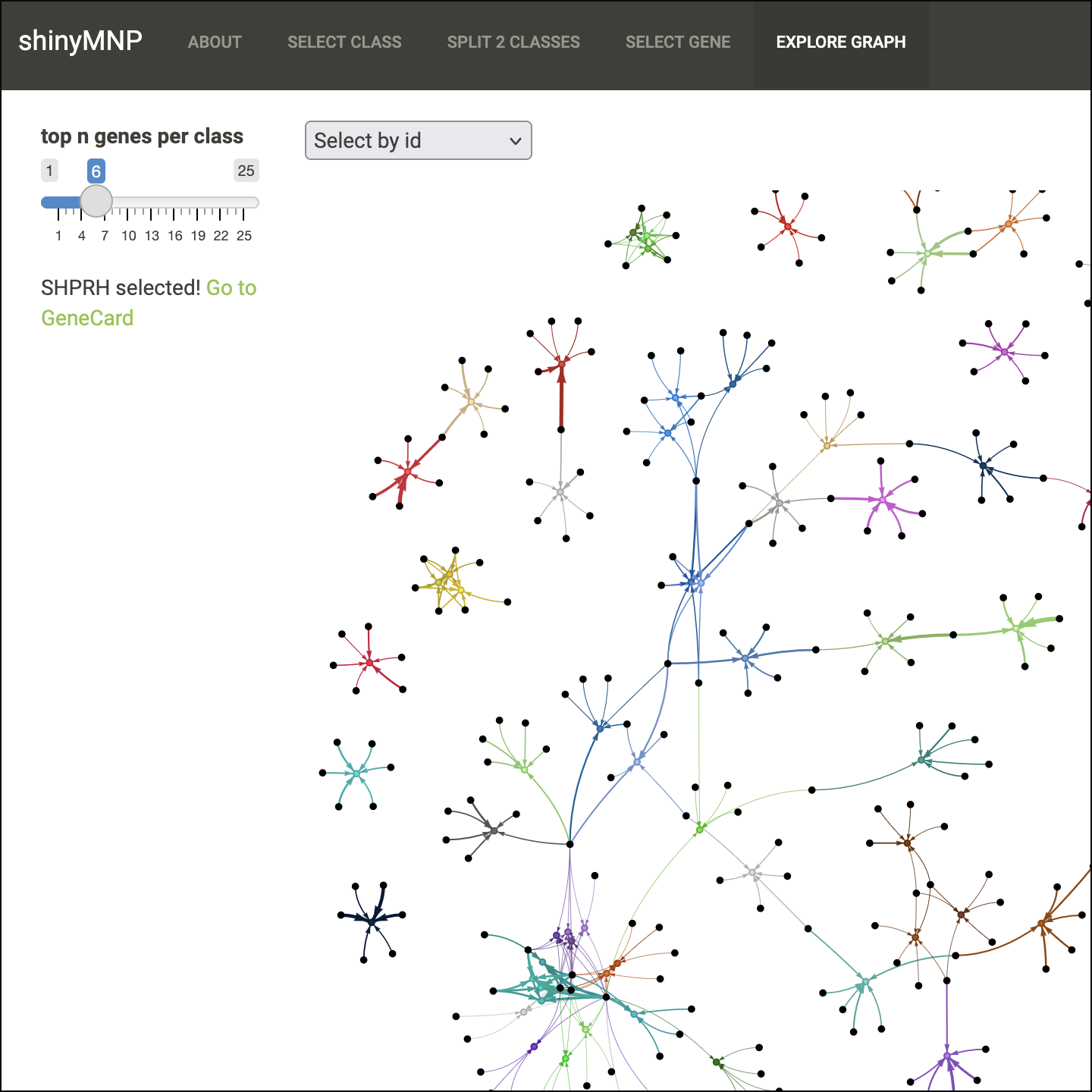
shinyMNP is an interactive web application to investigate predictive DNA methylation features underlying machine learning-based central nervous system tumor classifications. Different tabs allow for assessing most predictive features for each tumor class, most distinctive features between two tumor classes, and associated features with a gene of interest. For more information please see the associated publication.
https://hovestadtlab.shinyapps.io/shinyMNP/DNA methylation-based neuropathology
Web implementation of our DNA methylation-based classification system for central nervous system tumors. This classification system uses machine learning to turn patient-derived profiles into an accurate diagnosis. It is freely available and has become routine clinical assay that has been applied in tens-of-thousands of patients around the world.
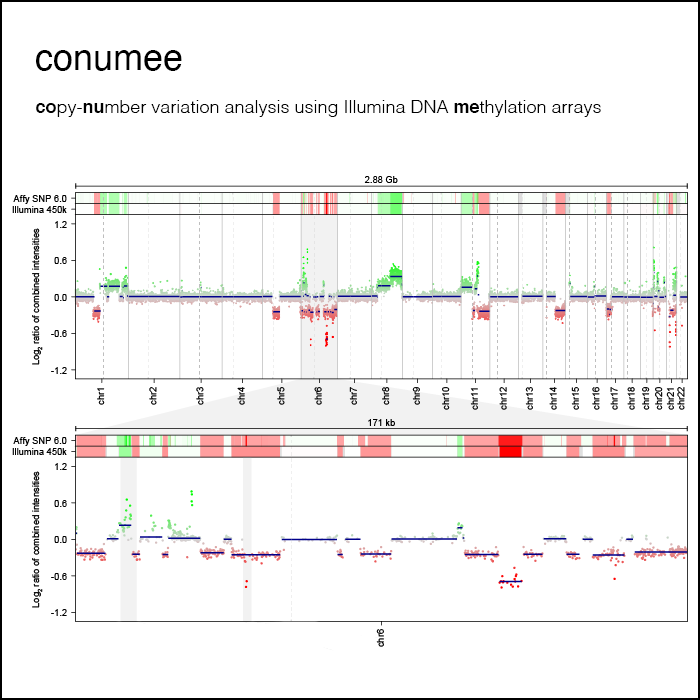
https://app.epignostix.com/Copy-number variation analysis from methylation arrays
The conumee package enables “off-label” copy-number variation calling from Illumina DNA methylation arrays. This added information is of importance for the classification of many types of brain tumors and replaces additional, often costly and laborious assays. The original version of conumee is available on Bioconductor. Conumee 2.0 is available on GitHub.
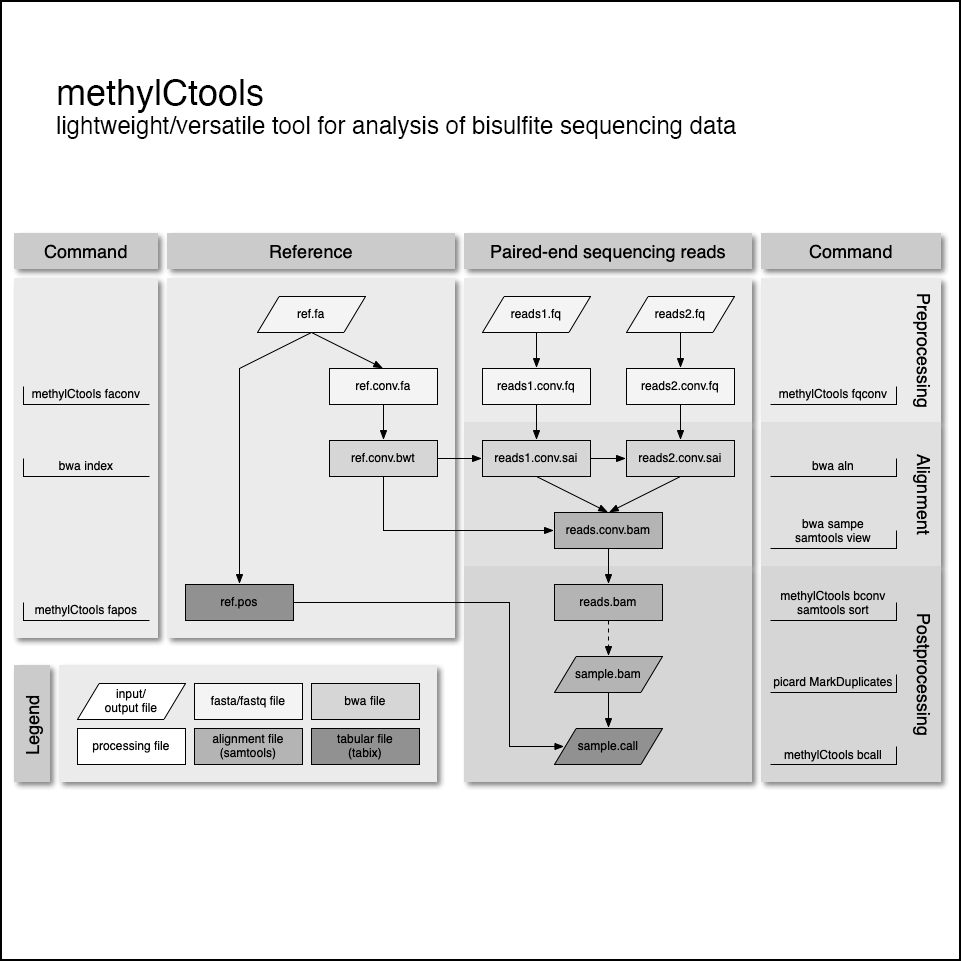
https://github.com/hovestadtlab/conumee2Anaysis of bisulfite sequencing data
methylCtools is a lightweight and versatile tool for the analysis of bisulfite-converted sequencing data. Bisulfite conversion is the gold-standard for quantifying CpG methylation and single basepair resolution. methylCtools wraps around popular tools like BWA and Samtools to provide both speed and functionality.
https://github.com/hovestadt/methylCtoolsSingle-cell transcriptome datasets
All our published single-cell transcriptome datasets are available through GEO to enable secondary analyses. Please reach out with any questions or comments.
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE119926