Publications
Please see Google Scholar for a complete list.
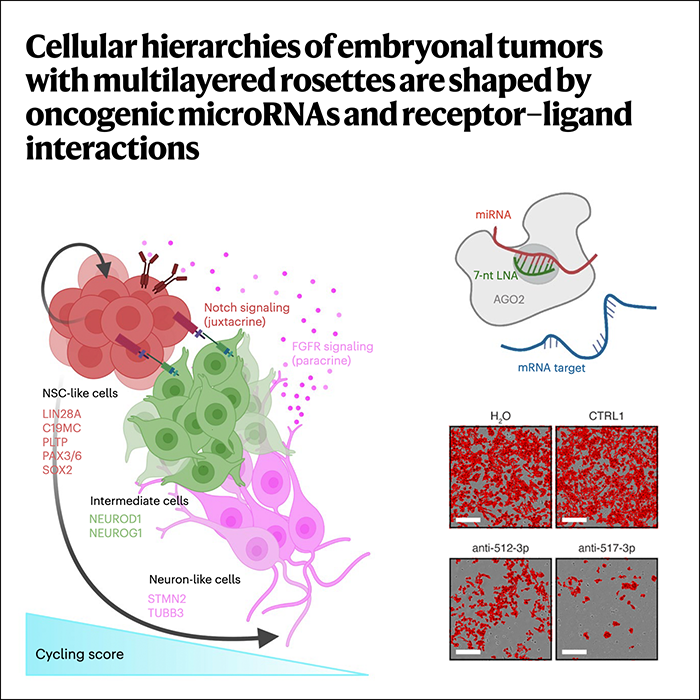
Cellular hierarchies of embryonal tumors with multilayered rosettes are shaped by oncogenic microRNAs and receptor–ligand interactions
Nature Cancer, 2025Beck A*, Gabler-Pamer L*, Cruzeiro GAV, Lambo S, …, Gojo J, Kool M†, Hovestadt V†, Filbin MG†.
We analyze Embryonal tumor with multilayered rosettes (ETMR), a pediatric brain tumor with dismal prognosis, using single-cell transcriptomics and multiplexed spatial imaging. We reveal a spatially distinct cellular hierarchy that spans highly proliferative neural stem-like cells and more differentiated neuron-like cells. C19MC is predominantly expressed in stem-like cells and controls a transcriptional network governing stemness and lineage commitment, as resolved by genome-wide analysis of microRNA-mRNA binding. Systematic analysis of receptor-ligand interactions between malignant cell types reveals fibroblast growth factor receptor and Notch signaling as oncogenic pathways that can be successfully targeted in preclinical models and in one patient with ETMR. Our study provides fundamental insights into ETMR pathobiology and a powerful rationale for more effective targeted therapies.
PubMed • Journal • PDF •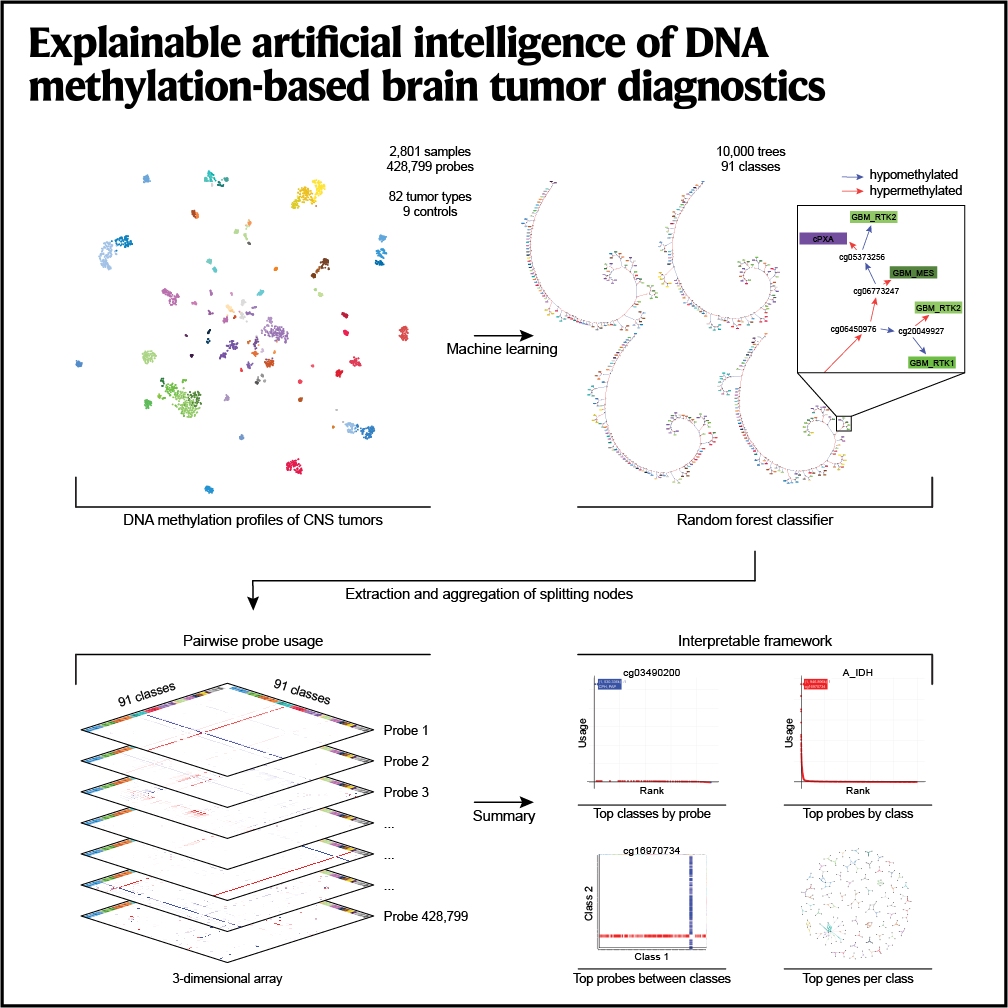
Explainable artificial intelligence of DNA methylation-based brain tumor diagnostics
Nature Communications, 2025Benfatto S, Sill M, Jones DTW, Pfister SM, Sahm F, von Deimling A, Capper D, Hovestadt V.
In this study we use explainable AI to explain the Heidelberg CNS tumor classifier. We show that DNA methylation probes in functional genomic regions of various sizes are predominantly employed to distinguish between different tumor classes, ranging from enhancers and CpG islands to large-scale heterochromatic domains. We detect a high degree of genomic redundancy, with many genes distinguishing individual tumor classes, explaining the robustness of the classifier and revealing potential targets for further therapeutic investigation.We anticipate that our resource will build up trust in machine learning in clinical settings, foster biomarker discovery and development of compact point-of-care assays, and enable further epigenome research of brain tumors.
PubMed • Journal • PDF •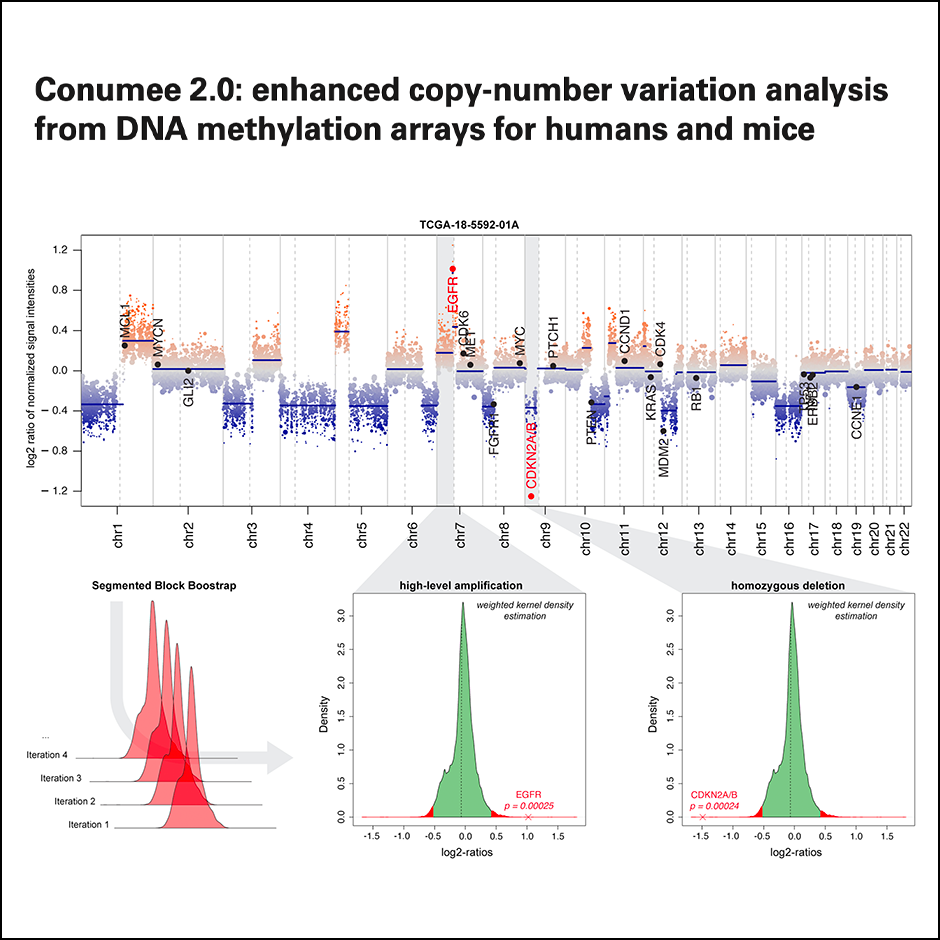
Conumee 2.0: Enhanced copy-number variation analysis from DNA methylation arrays for humans and mice
Bioinformatics, 2024Daenekas B, Perez E, Boniolo F, Stefan S, Benfatto S, Sill M, Sturm D, Jones DTW, Capper D, Zapatka M, Hovestadt V.
Detection of clinically relevant large and focal CNVs remains challenging when sample material or resources are limited. This has motivated us to create a software tool to infer CNVs from DNA methylation arrays which are often generated as part of clinical routines and in research settings. Conumee 2.0 combines tangent normalization, an adjustable genomic binning heuristic, and weighted circular binary segmentation to mitigate technical biases and batch effects. We further introduce a segmented block bootstrapping approach to detect focal alternations with high specificiay. Finally, our tool provides functionality to detect and summarize CNVs across large sample cohorts.
PubMed • Journal • PDF •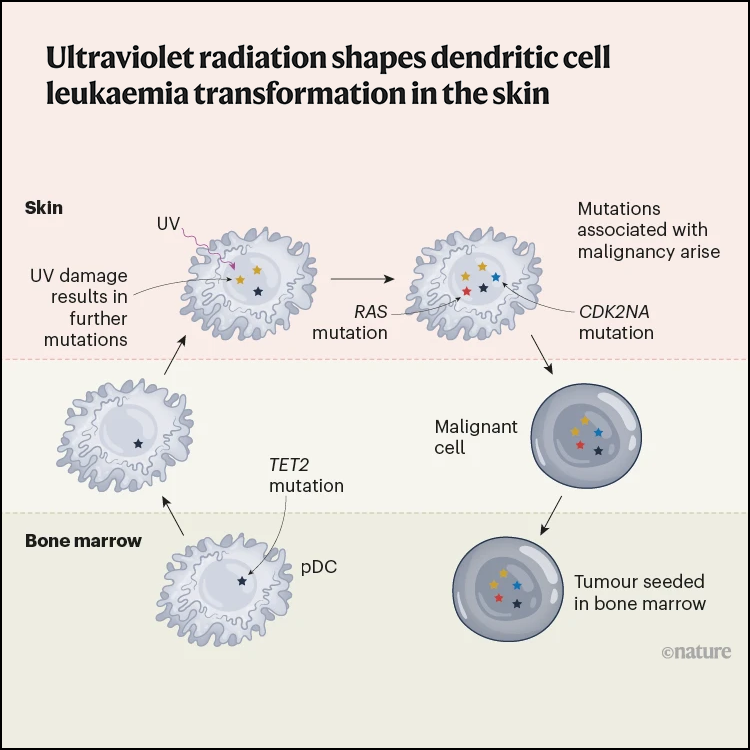
Ultraviolet radiation shapes dendritic cell leukaemia transformation in the skin
Nature, 2023Griffin GK, Booth CAG, Togami K, Chung SS, …, Bernstein BE, Hovestadt V†, van Galen P†, Lane AA†.
In this study we characterize blastic plasmacytoid dendritic cell neoplasm (BPDCN), a rare but curious type of acute leukemia that often presents with tumor cells in the skin. We perform longitudinal genomic/single-cell analyses of patient samples across multiple tissue sites. We find that BPDCN develops in a multi-step process across multiple tissue environments, from the bone marrow to the skin and back to the bone marrow. Exposure to UV radiation plays a key role in this process.
PubMed • Journal • PDF •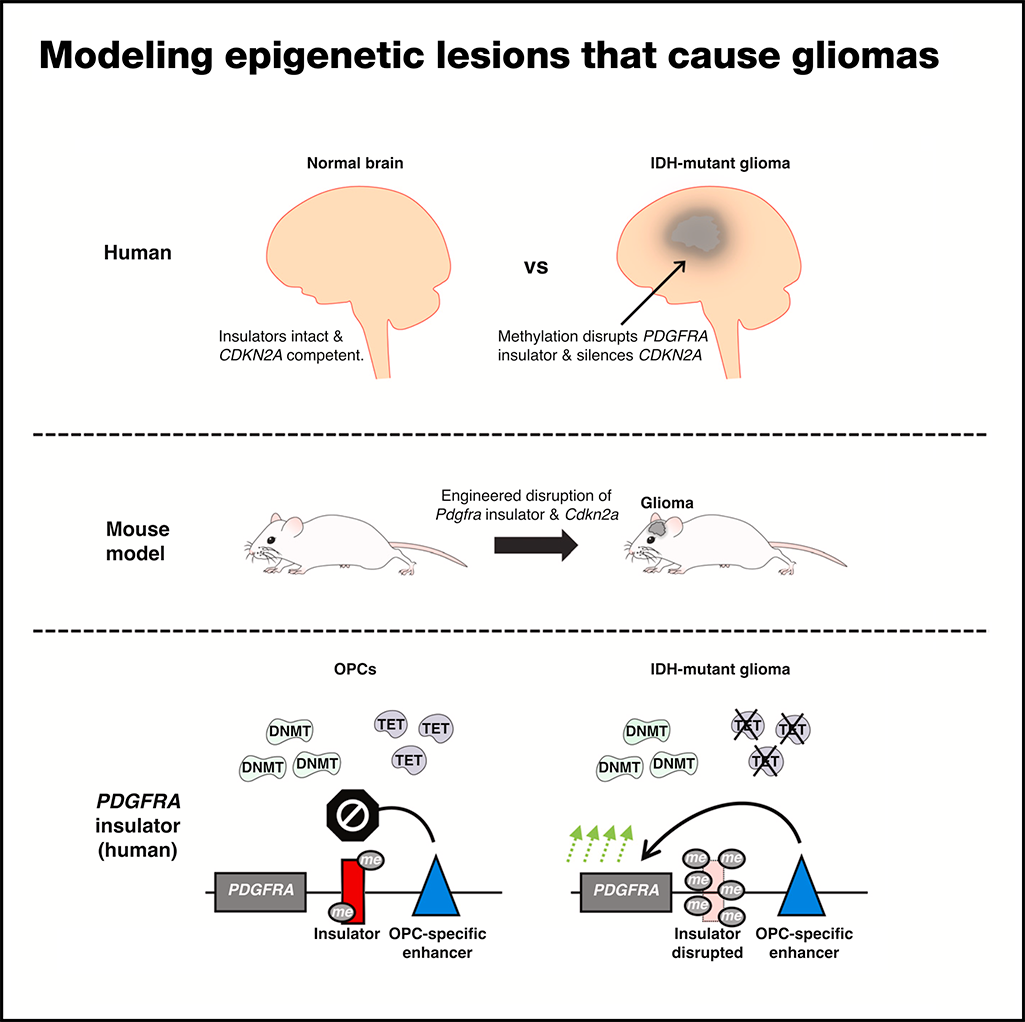
Modeling epigenetic lesions that cause gliomas
Cell, 2023Rahme GJ, Javed NM, Puorro KL, Xin S, Hovestadt V, Johnstone SE, Bernstein BE.
Epigenetic lesions that disrupt regulatory elements represent potential cancer drivers. Here, we model aberrations arising in IDH1-mutant gliomas, which exhibit DNA hypermethylation. We focus on a CTCF insulator near the PDGFRA oncogene that is recurrently disrupted by methylation in these tumors. We demonstrate that disruption of the syntenic insulator in mouse oligodendrocyte progenitor cells (OPCs) allows an OPC-specific enhancer to contact and induce Pdgfra, thereby increasing proliferation. Our study demonstrates the capacity of recurrent epigenetic lesions to drive OPC proliferation in vitro and gliomagenesis in vivo.
PubMed • Journal •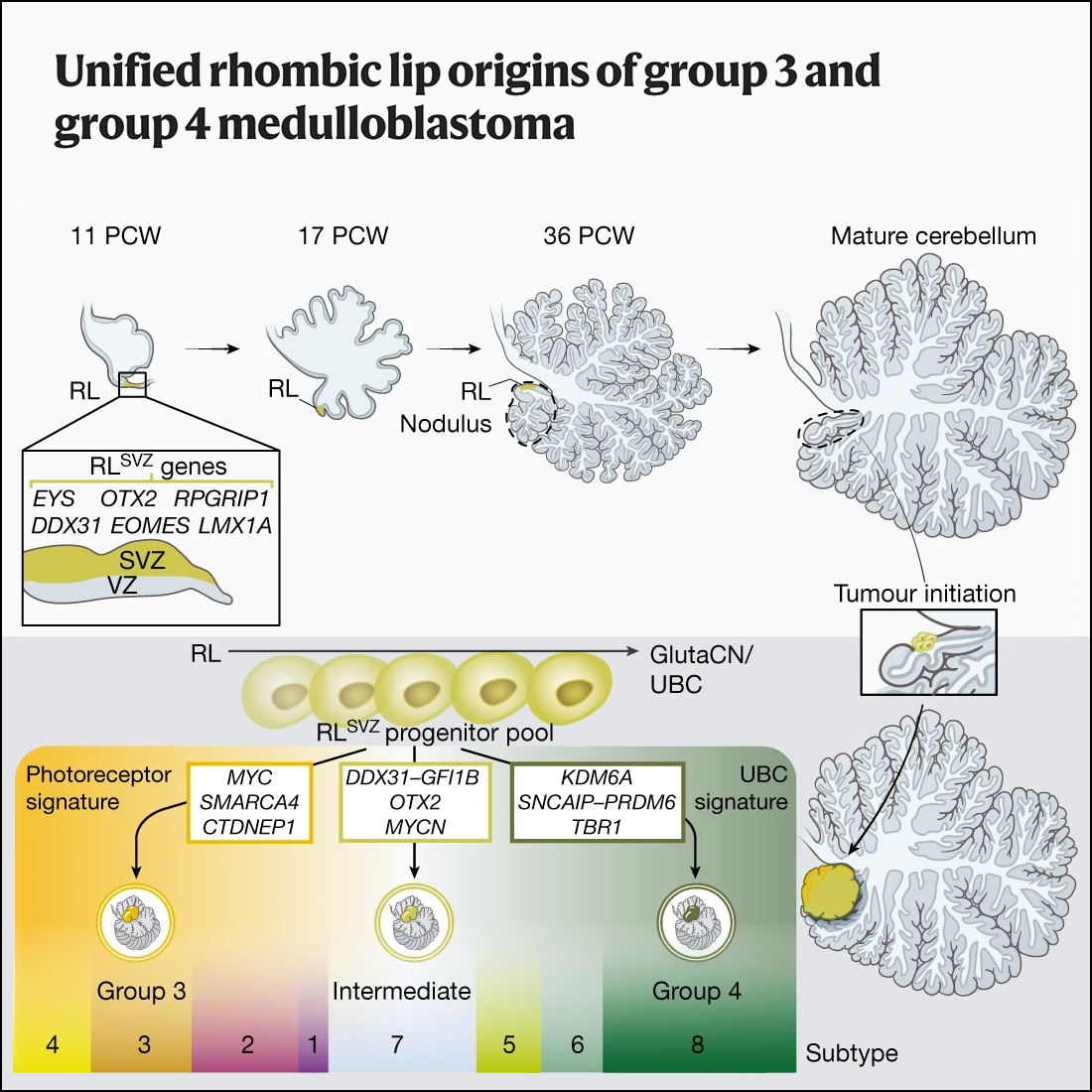
Unified rhombic lip origins of group 3 and group 4 medulloblastoma
Nature, 2022Smith KS*, Bihannic L*, Gudenas BL*, Haldipur P, …, Hovestadt V, Orr BA, Patay Z, Millen KJ†, Northcott PA†.
Here we use multi-omics to resolve the origins of medulloblastoma subgroups in the developing human cerebellum. Molecular signatures encoded within a human rhombic-lip-derived lineage trajectory aligned with photoreceptor and unipolar brush cell expression profiles that are maintained in group 3 and group 4 medulloblastoma, suggesting a convergent basis. Our results connect the molecular and phenotypic features of clinically challenging medulloblastoma subgroups to their unified beginnings in the rhombic lip in the early stages of human development.
PubMed • Journal • PDF •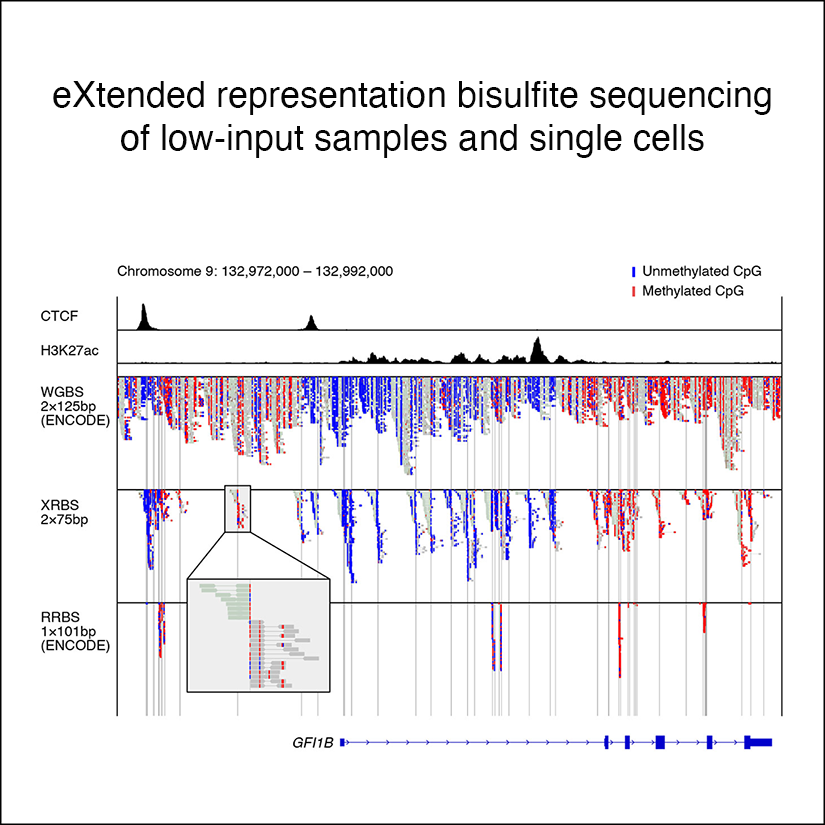
Extended-representation bisulfite sequencing of gene regulatory elements in multiplexed samples and single cells
Nature Biotechnology, 2021Shareef SJ, Bevill SM, Raman AT, Aryee MJ, van Galen P, Hovestadt V†, Bernstein BE†.
Here we present an extended-representation bisulfite sequencing (XRBS) method for targeted profiling of DNA methylation. Our design strikes a balance between expanding coverage of regulatory elements and reproducibly enriching informative CpG dinucleotides in promoters, enhancers and CTCF binding sites. Barcoded DNA fragments are pooled before bisulfite conversion, allowing multiplex processing and technical consistency in low-input samples.
PubMed • Journal • PDF •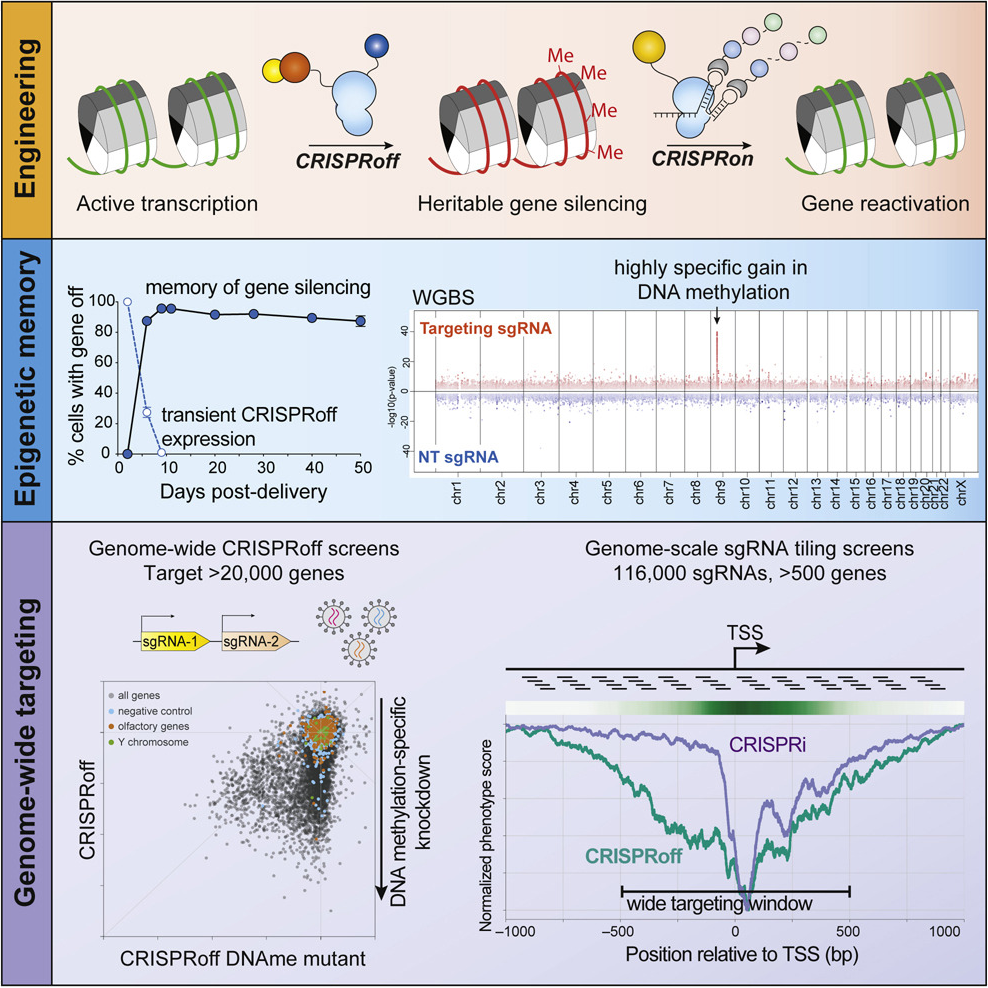
Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing
Cell, 2021Nunez JK, Chen J, Pommier GC, Cogan JZ, …, Bernstein BE, Hovestadt V, Gilbert LA†, Weissman JS†.
We present CRISPRoff, a programmable epigenetic memory writer consisting of a single dead Cas9 fusion protein that establishes DNA methylation and repressive histone modifications. Transient CRISPRoff expression initiates highly specific DNA methylation and gene repression that is maintained through cell division and differentiation of stem cells to neurons. The broad ability of CRISPRoff to initiate heritable gene silencing even outside of CGIs expands the canonical model of methylation-based silencing and enables diverse applications including genome-wide screens, multiplexed cell engineering, enhancer silencing, and mechanistic exploration of epigenetic inheritance.
PubMed • Journal • PDF •
Medulloblastomics revisited: biological and clinical insights from thousands of patients
Nature Reviews Cancer, 2020Hovestadt V, Ayrault O, Swartling FJ, Robinson GW, Pfister SM, Northcott PA.
This Review discusses recent genomic, epigenomic, transcriptomic and proteomic profiling studies of human medulloblastoma that have advanced our understanding of its disease subgroups. These efforts have provided new insights into the diverse biology of medulloblastomas that will hopefully lead to improved diagnosis and therapy.
PubMed • Journal • PDF •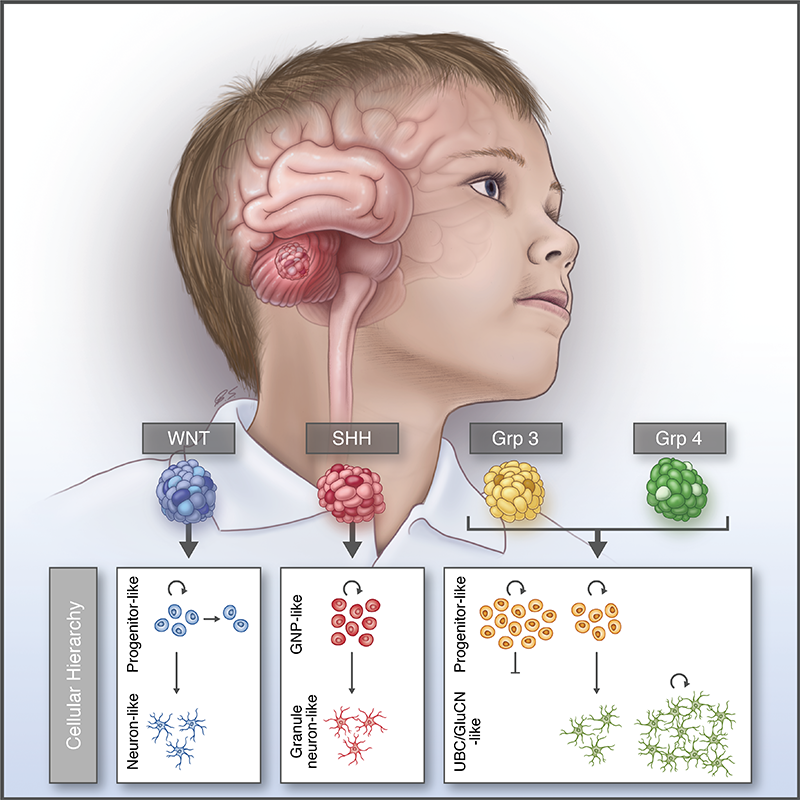
Resolving medulloblastoma cellular architecture by single-cell genomics
Nature, 2019Hovestadt V*, Smith KS*, Bihannic L*, Filbin MG*, …, Robinson GW, Bernstein BE†, Suvà ML†, Northcott PA†.
Characterization of medulloblastoma tissues using single-cell transcriptomics shows that the different molecular subtypes consist of distinct developmental phenotypes.
PubMed • Journal • PDF •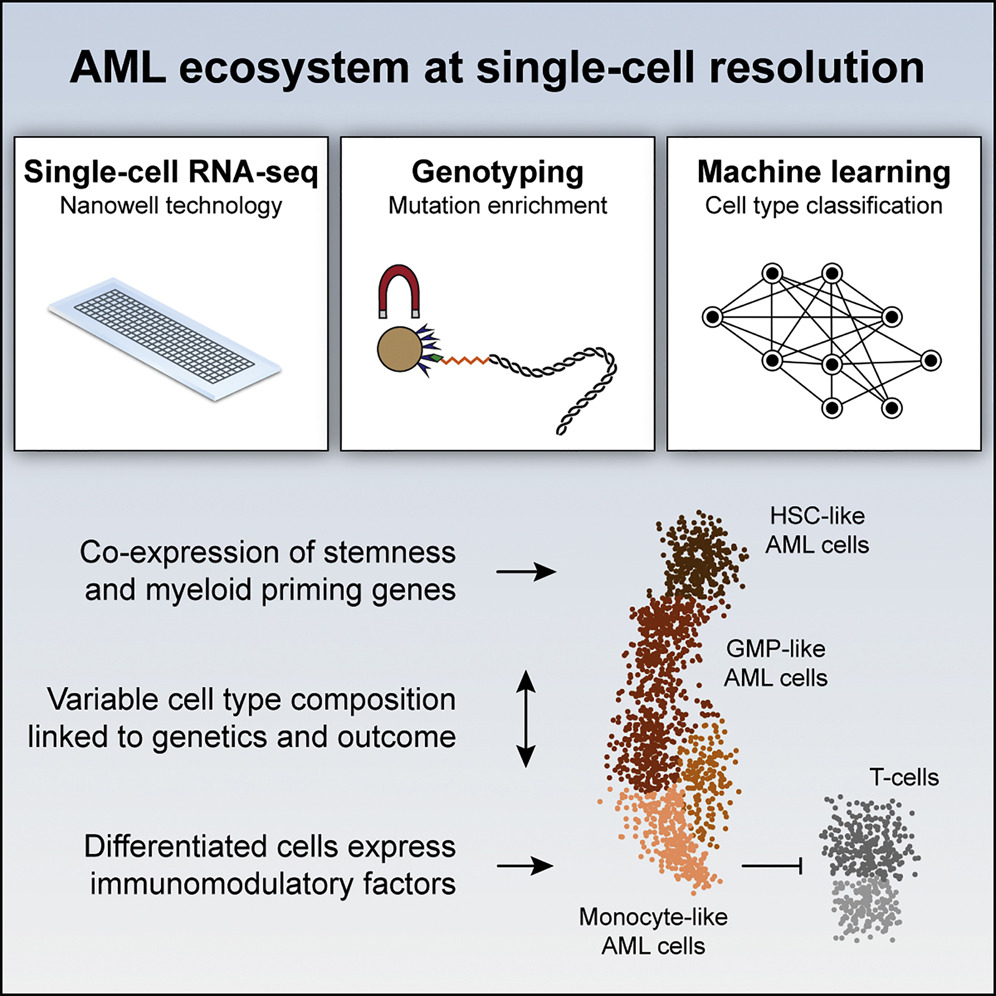
Single-Cell RNA-Seq Reveals AML Hierarchies Relevant to Disease Progression and Immunity
Cell, 2019van Galen P*, Hovestadt V*, Wadsworth II MH, Hughes TK, …, Shalek AK, Aster JC, Lane AA, Bernstein BE.
A combination of transcriptomics and mutational analyses in single cells from acute myeloid leukemia patients reveals the existence of distinct functional subsets and their associated drivers.
PubMed • Journal • PDF •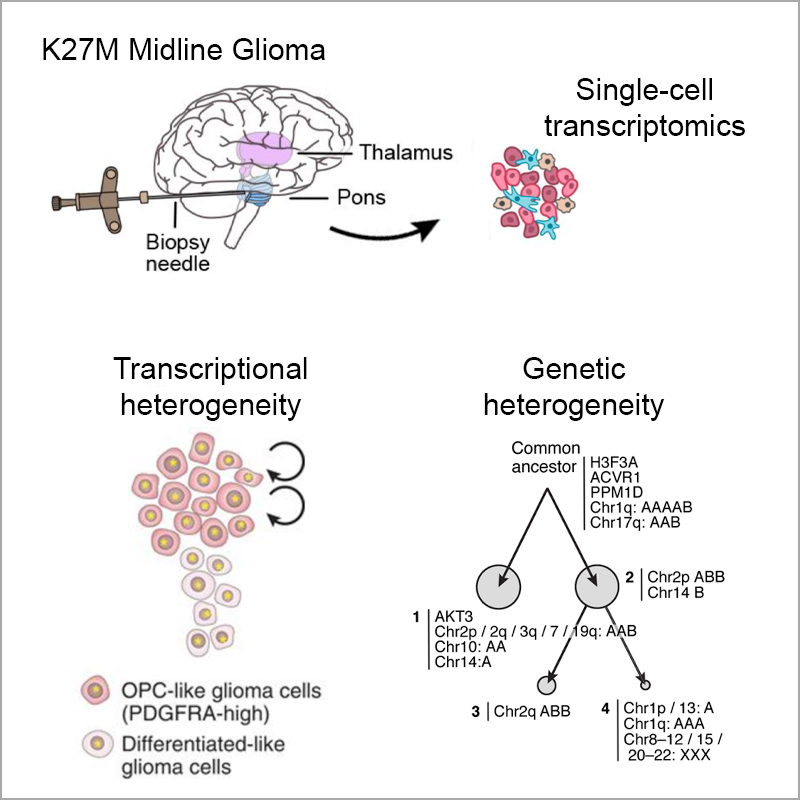
Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq
Science, 2018Filbin MG*, Tirosh I*, Hovestadt V*, Shaw ML, …, Golub TR, Regev A†, Bernstein BE†, Suvà ML†.
Diffuse midline gliomas with histone H3 lysine27-to-methionine mutations (H3K27M-glioma) are an aggressive type of childhood cancer with few options for treatment. Filbin et al. used a single-cell sequencing approach to study the oncogenic programs, genetics, and cellular hierarchies of H3K27M-glioma. Tumors were mainly composed of cells resembling oligodendrocyte precursor cells, whereas differentiated malignant cells were a smaller fraction. In comparison with other gliomas, these cancers had distinct oncogenic programs and stem cell–like profiles that contributed to their stable tumor-propagating potential. The analysis also identified a lineage-specific marker that may be useful in developing therapies.
PubMed • Journal • PDF •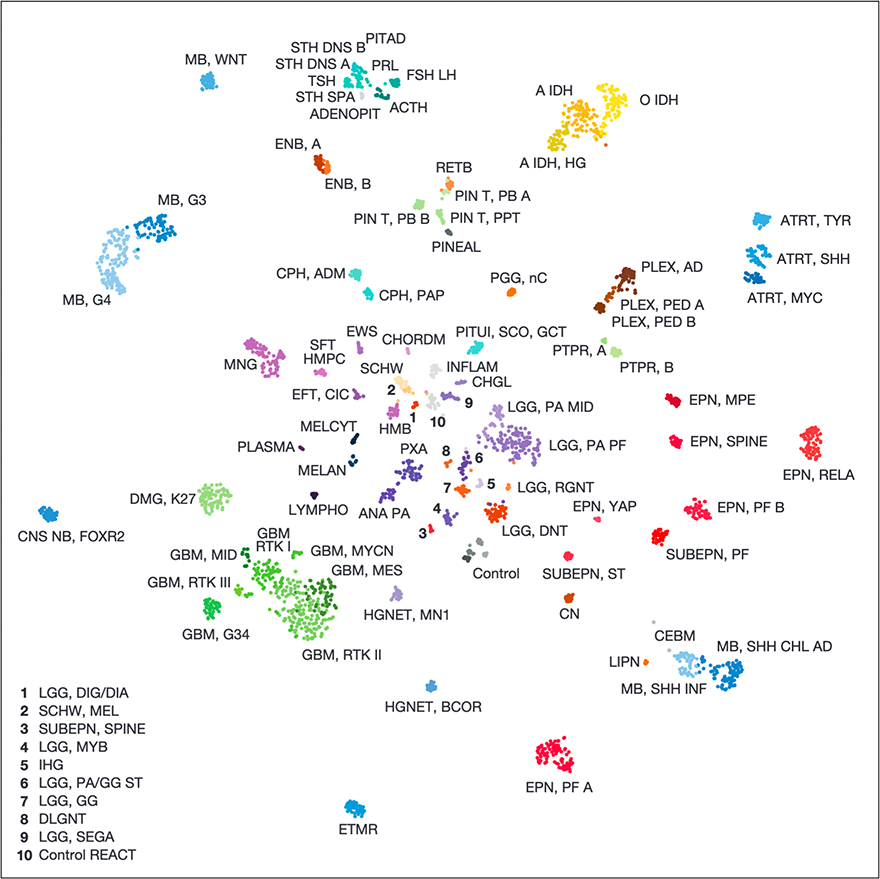
DNA methylation-based classification of central nervous system tumours
Nature, 2018Capper D*, Jones DTW*, Sill M*, Hovestadt V*, …, Brandner S, Korshunov A, von Deimling A†, Pfister SM†.
Precise cancer diagnoses are essential to ensure the best treatment plans for patients, but standardization of the diagnostic process has been challenging. The authors present a comprehensive approach for DNA-methylation-based classification of brain tumours. The tool improves diagnostic precision of standard methods, and is made available online for broad accessibility. The results illustrate the potential applications of molecular diagnosis tools.
PubMed • Journal • PDF •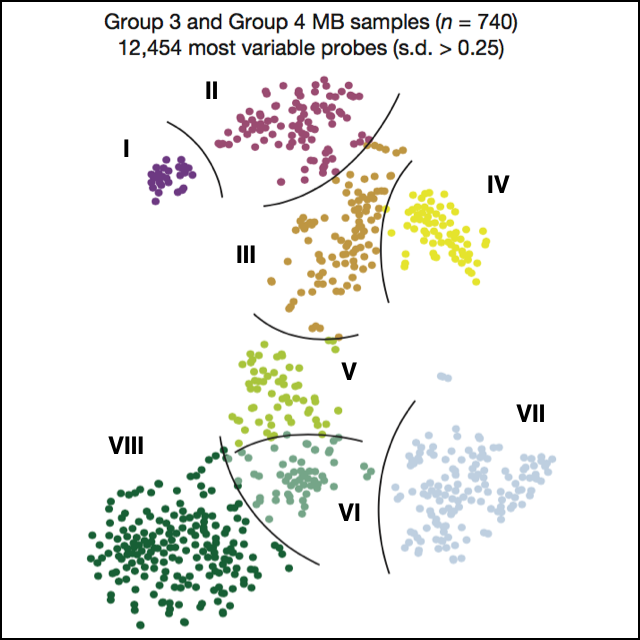
The whole-genome landscape of medulloblastoma subtypes
Nature, 2017Northcott PA*, Buchhalter I*, Morrissy AS*, Hovestadt V, …, Marra MA†, Pfister SM†, Taylor MD†, Lichter P†.
Medulloblastomas are highly malignant brain tumours that develop during childhood. Paul Northcott and colleagues analysed the whole-genome sequences of 491 medulloblastomas in order to characterize the genomic landscape across tumours and identify new drivers and mutational signatures. Their integrative genomic analyses, including methylation profiling of 1,256 medulloblastomas, identifies subgroup-specific driver mutations and suggests additional tumour subtypes. The authors assign driver mutations to a high proportion of the less well characterized Group 3 and Group 4, which together contribute to more than 60% of all medulloblastomas.
PubMed • Journal • PDF •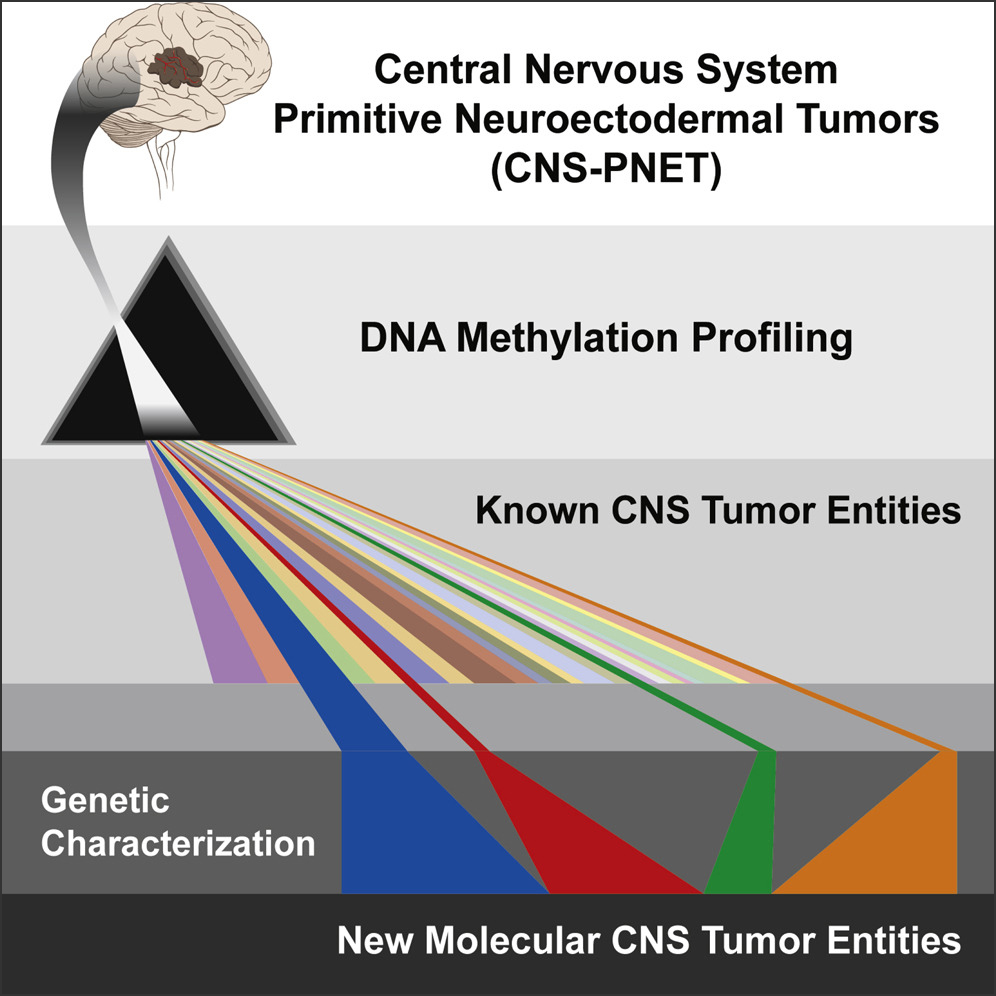
New Brain Tumor Entities Emerge from Molecular Classification of CNS-PNETs
Cell, 2016Sturm D*, Orr BA*, Toprak UH*, Hovestadt V*, …, Pfister SM, Ellison DW†, Korshunov A†, Kool M†.
Highly malignant primitive neuroectodermal tumors of the CNS (CNS-PNETs) have been challenging to diagnose and distinguish from other kinds of brain tumors, but molecular profiling now reveals that these cancers can be readily classified into some known tumor types and four new entities with distinct histopathological and clinical features, paving the way for meaningful clinical trials.
PubMed • Journal • PDF •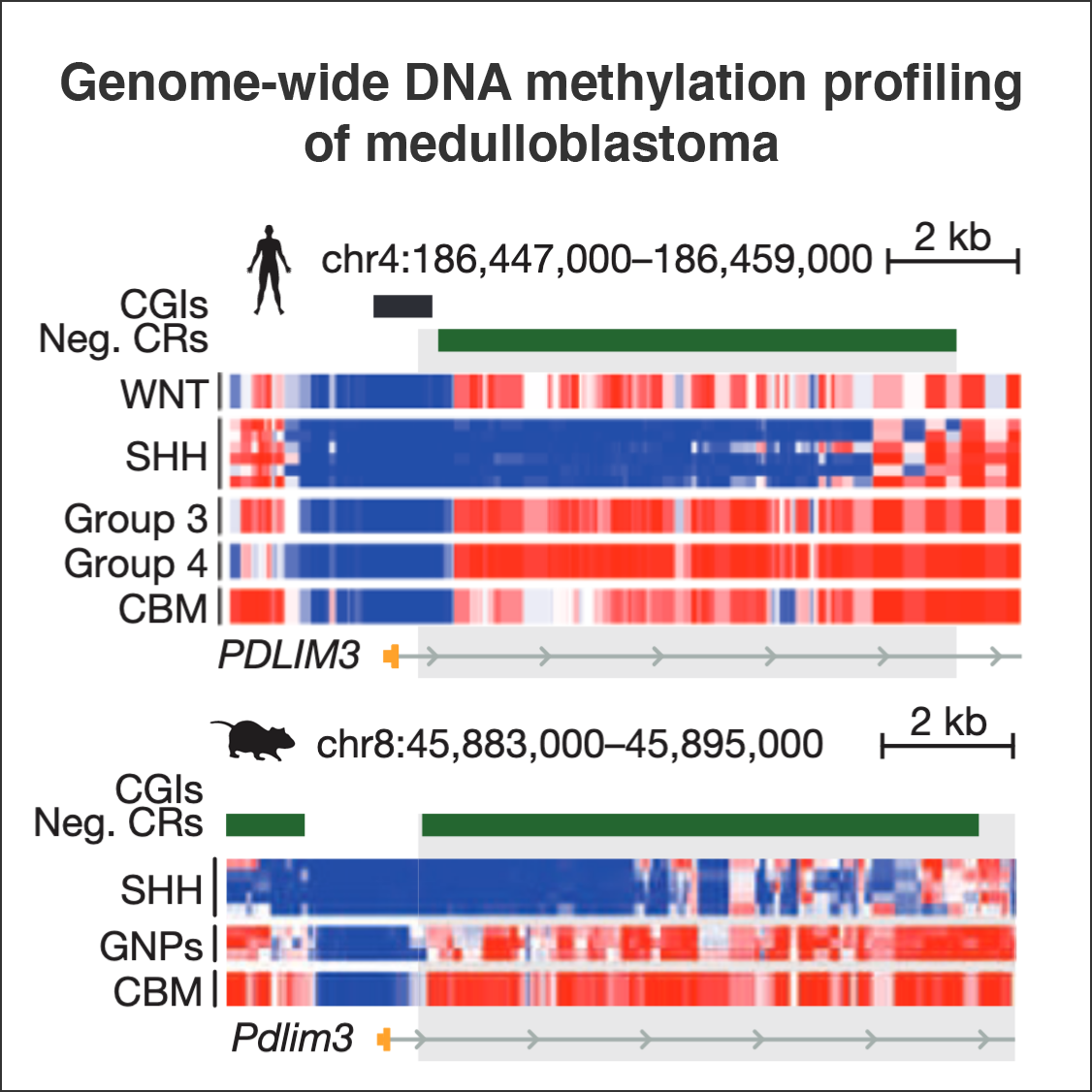
Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing
Nature, 2014Hovestadt V*, Jones DTW*, Picelli S, Wang W, …, Zapatka M, Radlwimmer B†, Pfister SM†, Lichter P†.
Medulloblastoma is the most common malignant childhood brain tumour. Here, the authors present a comprehensive genome-wide DNA methylation data set from human and mouse tumours, coupled with analysis of histone modifications, RNA transcripts and genome sequencing. This integrative analysis uncovers a wealth of alterations in the methylome and should help identify new targets for potential therapeutic intervention.
PubMed • Journal • PDF •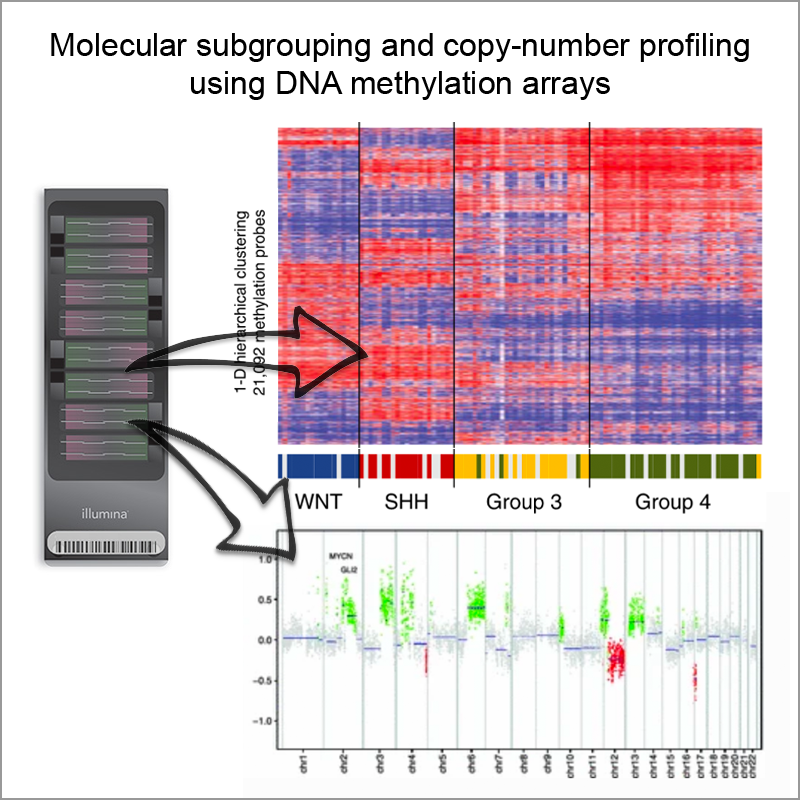
Robust molecular subgrouping and copy-number profiling of medulloblastoma from small amounts of archival tumour material using high-density DNA methylation arrays
Acta Neuropathologica, 2013Hovestadt V, Remke M, Kool M, Pietsch T, …, Lichter P, Taylor MD, Pfister SM, Jones DTW
We demonstrate here a method for reliable classification of medulloblastoma into molecular subgroups, and tumour copy-number profiling, using a commercially available DNA methylation array platform that performs well on either frozen or FFPE tumour material. We believe that this platform holds great potential for refining the information obtainable from large, archival tumour series. Most importantly, we also expect that this will become one of the key technologies for risk stratification and patient cohort selection in the next generation of large, biology-led, multi-centre clinical trials.
PubMed • Journal • PDF •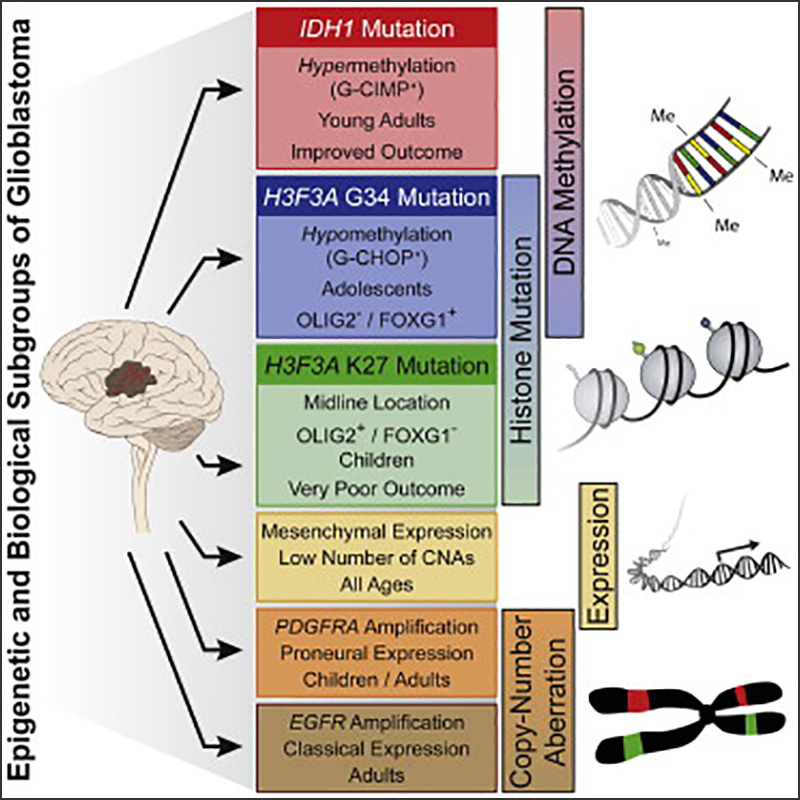
Hotspot Mutations in H3F3A and IDH1 Define Distinct Epigenetic and Biological Subgroups of Glioblastoma
Cancer Cell, 2012Sturm D*, Witt H*, Hovestadt V*, Khuong-Quang DA*, …, Lichter P, Plass C†, Jabado N†, Pfister SM†
Glioblastoma (GBM) is a brain tumor that carries a dismal prognosis and displays considerable heterogeneity. We have recently identified recurrent H3F3A mutations affecting two critical amino acids (K27 and G34) of histone H3.3 in one-third of pediatric GBM. Here, we show that each H3F3A mutation defines an epigenetic subgroup of GBM with a distinct global methylation pattern, and that they are mutually exclusive with IDH1 mutations, which characterize a third mutation-defined subgroup. Three further epigenetic subgroups were enriched for hallmark genetic events of adult GBM and/or established transcriptomic signatures.
PubMed • Journal • PDF •